Request a quote
| Appearance : | White amorphous powder, lyophilized | |
|---|---|---|
| Activity : | GradeⅢ 400 U/mg-solid or more (NAD+) | |
| Stability : | Stable at −20 ℃ for at least one year | |
|---|---|---|
| Stable at 5 ℃ for at least 6 months (liquid form) | ||
| Molecular weight : | 104,000(two subunits of approx. 55,000) 1,2) | |
| Appearance | White amorphous powder, lyophilized | |
|---|---|---|
| Activity | GradeⅢ 400 U/mg-solid or more (NAD+) | |
| Contaminants | Creatine phosphokinase | ≤ 1×10-3 % |
| Phosphoglucomutase | ≤ 1×10-3 % | |
| 6-Phosphogluconate dehydrogenase | ≤ 5×10-3 % | |
| Phosphoglucose isomerase | ≤ 1×10-2 % | |
| Glutathione reductase | ≤ 1×10-3 % | |
| Hexokinase | ≤ 1×10-2 % | |
| Myokinase | ≤ 1×10-2 % | |
| NADH oxidase | ≤ 1×10-2 % | |
| NADPH oxidase | ≤ 1×10-2 % | |
| Stability | Stable at −20 ℃ for at least one year(Fig.1) | |
|---|---|---|
| Stable at 5 ℃ for at least 6 months (liquid form)(Fig.3) | ||
| Molecular weight | 104,000(two subunits of approx. 55,000) 1,2) | |
| Isoelectric point | 4.6 2) | |
| Michaelis constants 2) | NAD+ linked | 1.06×10-4 M (NAD+), 5.27×10-5 M (G-6-P) |
| NADP+ linked | 5.69×10-6 M (NADP+), 8.1×10-5 M (G-6-P) | |
| Structure | Neither cysteine nor cystine residues is present in the enzyme molecule 1) and essential lysine is indicated to be at active site. 3) | |
| Inhibitors | Acyl-CoA,4) ATP,4) mental ions etc. (Table 1) | |
| Optimum pH | 7.8(Fig.4) | |
| Optimum temperature | 50 ℃(Fig.5) | |
| pH Stability | pH 5.5−7.5 (30 ℃, 17 hr)(Fig.6) | |
| Thermal stability | below 37 ℃ (pH 8.0, 30 min)(Fig.7) | |
| Substrate specificity | Either NAD+ or NADP+ serves as coenzyme, the reaction velocity with NAD+ being approximately 1.8 times greater than with NADP.+5) DGlucose-6-phosphate is a preferential substrate for the enzyme, although D-glucose reacts slowly.6) Fructose-6-phosphate, fructose1, 6-diphoshate and ribose-5-phosphate are not considered to be substrates.7) | |
This enzyme is useful for enzymatic determation of NAD+(NADP+) and G-6-P, and the activities of phosphoglucose isomerase, phosphoglucomutase and hexokinase. This enzyme is also used for enzymatic determination of glucose in combination with hexokinase (HXK-311).
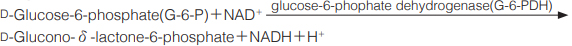
The formation of NADH is measured at 340 nm by spectrophotometry.
One unit causes the formation of one micromole of NADH per minute under the conditions detailed below.
| A. Tris-HCl buffer, pH 7.8 | 55 mM (containing 3.3 mM magnesium chloride) | |
|---|---|---|
| B. NAD+ solution | 60 mM (Should be prepared fresh) | |
| C. G-6-P solution | 0.1 M glucose-6-phosphate (should be prepared fresh) | |
| D. Enzyme diluent | 5 mM Tris-HCl buffer, pH 7.5, containing 0.1 % of bovine serum albumin. | |
1.Prepare the following reaction mixture in a cuvette (d = 1.0 cm) and equilibrate at 30 ℃ for about 5 minutes.
| 2.7 mL | Tris-HCl buffer, pH 7.8 | (A) |
| 0.1 mL | NAD+ solution | (B) |
| 0.1 mL | G・6・P solution | (C) |
| Concentration in assay mixture | |
|---|---|
| Tris-HCl buffer | 50 mM |
| G-6-P | 3.3 mM |
| NAD+ | 2.0 mM |
| MgCl2 | 3.0 mM |
| BSA | 33 μg/mL |
2.Add 0.1 mL of the enzyme solution* and mix by gentle inversion.
3.Record the increase in optical density at 340 nm against water for 4 to 5 minutes with a spectrophotometer thermostated at 30 ℃ and calculate the ΔOD per minute from the initial linear portion of the curve (ΔOD test).
At the same time, measure the blank rate (ΔOD blank) using the same method as the test except that the enzyme diluent is added instead of the enzyme solution.
*Dissolve the enzyme preparation in ice-cold enzyme diluent (D) and dilute to 0.05−0.20 U/mL with the same buffer, immediately before the assay.
Activity can be calculated by using the following formula :
Volume activity (U/mL) =
ΔOD/min (ΔOD test−ΔOD blank)×Vt×df
6.22×1.0×Vs
= ΔOD/min×4.82×df
Weight activity (U/mg) = (U/mL)×1/C
| Vt | : Total volume (3.0 mL) |
| Vs | : Sample volume (0.1 mL) |
| 6.22 | : Millimolar extinction coefficient of NADH (cm2/micromole) |
| 1.0 | : Light path length (cm) |
| df | : Dilution facter |
| C | : Enzyme concentration in dissolution (c mg/mL) |
1)A.Ishaque,M.Mihausen and H.R.Levy; Biochem. Biophys. Res. Comm., 59, 894 (1974).
2)C. Olive, M.E. Geroch and H.R.Levy; J.Biol.Chem., 246, 2043 (1971).
3)M.Milhausen and H.R. Levy; Eur.J.Biochem., 50, 453 (1975).
4)E.L.Coe and L.-H.Hsu; Biochem. Biophys. Res. Comm., 53, 66 (1973).
5)C.Olive and H.R. Levy; Biochem., 6, 730 (1967).
6)R.P.Metzger, S.A. Metzger and R.L. Parsons; Arch Biochem. Biophys., 149, 102 (1972).
7)Methods in Enzymology, Vol, 1, p328 (S.P.Colowick and N.O.Kapalan,eds.), Academic Press, New York (1955).
[The enzyme dissolved in 50 mM Tris-HCl buffer,pH 7.5 (5.25 U/mL) was incubated with each chemcal for 1 hr at 30 ℃.]
| Chemical | Concn.(mM) | Residual activity(%) |
|---|---|---|
| None | – | 100 |
| Metal salt | 2.0 | |
| AgNO3 | 86 | |
| Ba(OAc)2 | 51 | |
| CaCl2 | 90 | |
| Cd(OAc)2 | 74 | |
| CoCl2 | 80 | |
| CuSO4 | 66 | |
| FeCl3 | 0 | |
| FeSO4 | 1 | |
| HgCl2 | 84 | |
| MgCl2 | 90 | |
| MnCl2 | 89 | |
| NiCl2 | 89 | |
| Pb(OAc)2 | 3 | |
| Zn(OAc)2 | 67 | |
| ZnSO4 | 53 | |
| KF | 2.0 | 93 |
| NaF | 20.0 | 98 |
| NaN3 | 20.0 | 93 |
| Chemical | Concn.(mM) | Residual activity(%) |
|---|---|---|
| NEM | 2.0 | 91 |
| PCMB | 2.0 | 96 |
| MIA | 2.0 | 14 |
| Iodoacetamide | 2.0 | 0 |
| EDTA | 5.0 | 94 |
| (NH4)2SO4 | 20.0 | 98 |
| Borate | 20.0 | 95 |
| o-Phenanthroline | 2.0 | 93 |
| α,α′-Dipyridyl | 2.0 | 95 |
| Urea | 2.0 | 93 |
| Guanidine | 2.0 | 93 |
| Hydroxylamine | 2.0 | 91 |
| Na-cholate | 1.0 % | 102 |
| Triton X-100 | 1.0 % | 100 |
| Brij 35 | 1.0 % | 4 |
| SDS | 0.1 % | 0 |
| Tween 20 | 0.1 % | 101 |
| Span 20 | 0.1 % | 99 |
| DAC | 0.1 % | 0 |
Ac, CH3CO; NEM, N-Ethylmaleimide; PCMB, p-Chloromercuribenzoate; MIA, Monoiodoacetate; EDTA, Ethylenediaminetetraacetate; SDS, Sodium dodecyl sulfate; DAC, Dimethylbenzylalkylammoniumchloride.
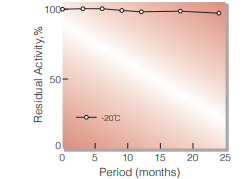
Fig.1. Stability (Powder form)
(kept under dry conditions)
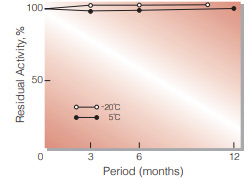
Fig.2. Stability (Powder form)
(kept under dry conditions)
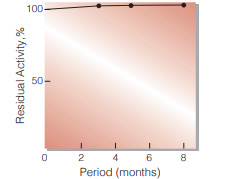
Fig.3. Stability (Liquid form at 5 ℃)
enzyme concentration:5,000 U/mL composition:3.2 M ammonium sulfate,pH 6.0
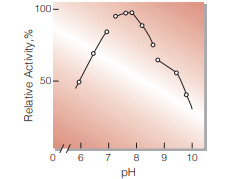
Fig.4. pH-Activity
30 ℃ in the following buffer solution: pH 5.7-6.8, 15 mM Veronal-CH3COONaHCI;pH 6.8-8.5,50 mM Tris-HCI; pH 8.5-9.5, 50 mM glycine-NaOH
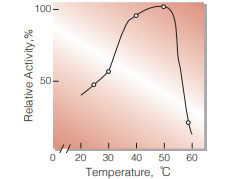
Fig.5. Temperature activity
(in 50 mM Tris-HCI buffer, pH 7.8)
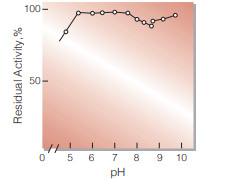
Fig.6. pH-Stability
30 ℃, 17 hr-treatment with the following buffer solution: pH 5.0-7.8, 30 mM VeronalCH3COONa-HCI;pH 7.5-8.5, 0.1 M Tris-HCI; pH 8.5-9.5,0.1 M glycine-NaOH
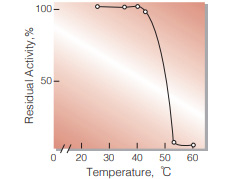
Fig.7. Thermal stability
30 min-treatment with 5.0 mM glycineNaOH buffer, pH 8.0, containing 0.1 % of bovine serum albumin
| Size | 1 MG, 10 MG, 5 MG |
|---|
|
|
|
0% |
|
|
|
0% |
|
|
|
0% |
|
|
|
0% |
|
|
|
0% |
| Name | Galactokinase; BiGalK |
| Product Code | EN01018 |
| E.C. | 2.7.1.6 |
| Product Description | E. coli recombinant galactokinase from Bifidobacterium infantis |
| Unit Definition | One unit is defined as the amount of enzyme that catalyzes the formation of 1 μmol of Gal-1-P from galactose and ATP per minute at 37 °C. |
10 in stock
| Name | CMP-sialic acid synthetase; NmCSS |
| Product Code | EN01015 |
| E.C. | 2.7.7.43 |
| Product Description | E. coli recombinant α2,6sialyltransferase from Photobacterium damsel |
| Unit Definition | One unit is defined as the amount of enzyme that catalyzes the formation of 1 μmol CMP-Sia from sialic acid and CTP per minute at 37 °C. |
10 in stock
| Name | Endo-β-N-acetylglucosaminidase A;Endo-A |
| Product Code | EN01023 |
| E.C. | 3.2.1.96 |
| Product Description | E. coli recombinant endo-β-N-acetylglucosaminidase from Arthrobacter protophormia |
| Unit Definition | One unit is defined as the amount of enzyme that catalyzes the release of 1 nmol N-glycan from RNaseB per minute at 37 °C. |
10 in stock
| Name | α2,3-sialyltransferase; PmST1 |
| Catalog Number | EN01002 |
| E.C. | 2.4.99.4 |
| Product Description | E. coli recombinant α2,3-sialyltransferase from Pasteurella multocida (P-1059) |
| Unit Definition | One unit is defined as the amount of enzyme that catalyzes the formation of 1 µmol Siaα2,3Lac from CMP-Sia and lactose per minute at 37 °C. |
10 in stock
Not a member? Create an account
Already got an account? Sign in here
Reviews
There are no reviews yet.